piRNA Pathway (Group Stallmeyer)
Novel disease genes for male infertility: Impact of the testicular piRNA pathway and genetic causes beyond
The underlying causes of impaired spermatogenesis leading to male infertility are often elusive. However, genetic factors are considered to play a major role in the disease.
In our projects, we aim to identify and functionally characterise novel genetic causes of the disease, with a particular focus on genes thought to be involved in the piRNA pathway or located on the human sex chromosomes.
Identifying piRNA pathway linked monogenic causes of azoospermia
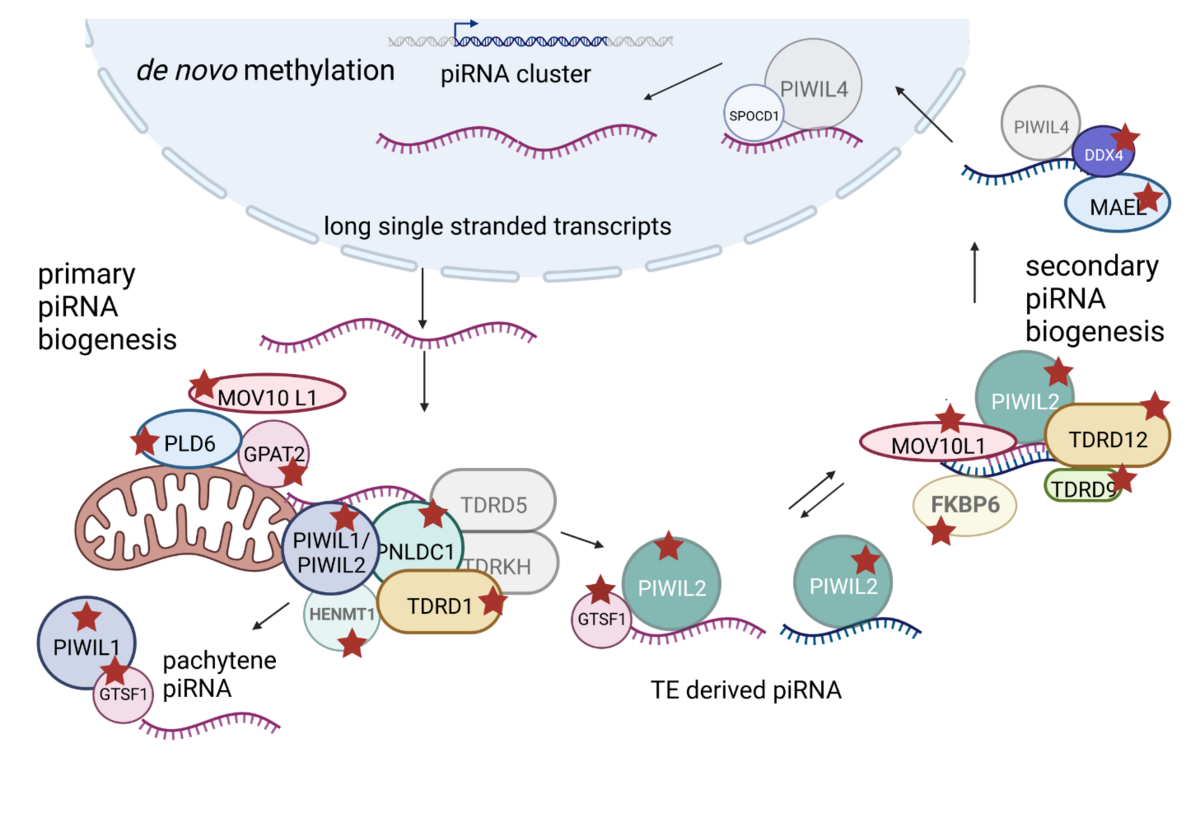
Little is known about the consequences of loss-of piRNA pathway gene function in men on spermatogenesis, and the precise expression pattern and subcellular localisation of the encoded proteins have not yet been systematically analysed. Therefore, we aim to identify infertile men with biallelic potentially pathogenic variants and analyse the consequences of the dysfunctionally encoded protein on piRNA generation as well as downstream piRNA mediated germ cell-specific functions.
Sex chromosomes-linked monogenic causes of azoospermia
The sex chromosomes (X and Y) are critical players in human male fertility since they contain several genes essential for sex determination and reproductive function. The X and Y-linked germ cell-specific genes are particularly attractive for the study of human male infertility, because, unlike autosomal recessive mutations, pathogenic variants in these genes would not be masked by a normal allele in males. On the human Y chromosome, despite the presence of the sex determination factor SRY, genes located in the AZF regions have been demonstrated to be essential for proper spermatogenesis. However, the key spermatogenic factors within these regions remain unclear. By screening the whole exome sequencing data of the MERGE cohort for rare loss-of function variants in genes with testis-specific expression, we aim to identify novel sex chromosomal genes associated with the disease.
As a first result, we demonstrated that DDX3Y is the key spermatogenic factor within the Y-chromosomal AZFa region.

In perspective, our projects will scrutinise the crucial piRNA pathway components and sex chromosomal infertility genes in men and thus further reduce the proportion of diagnostically unsolved male infertility cases. By in-depth functional characterisation of identified, potentially pathogenic variants, the project will also increase the knowledge of cellular functions of the respective disease genes and provide phenotype characterisation-dependent information on the patient’s chances to father a child via assisted reproduction techniques such as in vitro sperm extraction and intracytoplasmic injection.


